
Although still a young endeavour, tremendous advances have been made in ancient DNA research to type ancient genetic markers. Ever-refined approaches to extracting and amplifying DNA from organellar and nuclear sources have opened new insights and stimulated entirely new research questions about past human and animal populations.
In Africa, most efforts using ancient DNA have focused on human origins research and the history of animal domestication and pastoralism. Only a single sample of Nguni cattle has been examined in the study of the spread of African pastoralism. Our work will expand the available genetic data from modern and pre-colonial domestic livestock and help us understand herd diversity and management.
Our Research
In our research we are using mitochondrial DNA, or mtDNA, because there is a higher likelihood of recovering intact segments of DNA in mitochondrial organelles than with nuclear DNA. There is only one nucleus in a cell, but there are hundreds of mitochondria and chloroplasts. So, there are thousands of copies of mitochondrial or chloroplast DNA in any one cell even though they make up a small proportion of the total genomic DNA.
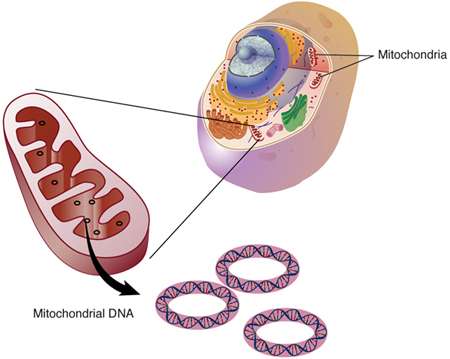
Our interests lay in how maternal cattle lineages can inform us about managing cattle as a form of wealth and as a source of meat, bone, hide, and sinew for food, tools, clothing, ceremonial equipment, and the shields and regalia for the Zulu army.
The inclusion of ancient DNA evidence in our project has three main purposes.
- In South Africa, complete mitochondrial genomes are only available from a single sample of Nguni cattle, which were sourced from five herds across eSwatini (Horsbourgh et al. 2013). Most belong to the T1b1 subhaplotype group. We will expand that sample to two further carefully managed herds, including the Zulu royal herd, in collaboration with members of the Nguni Cattle Breeders Society of South Africa.
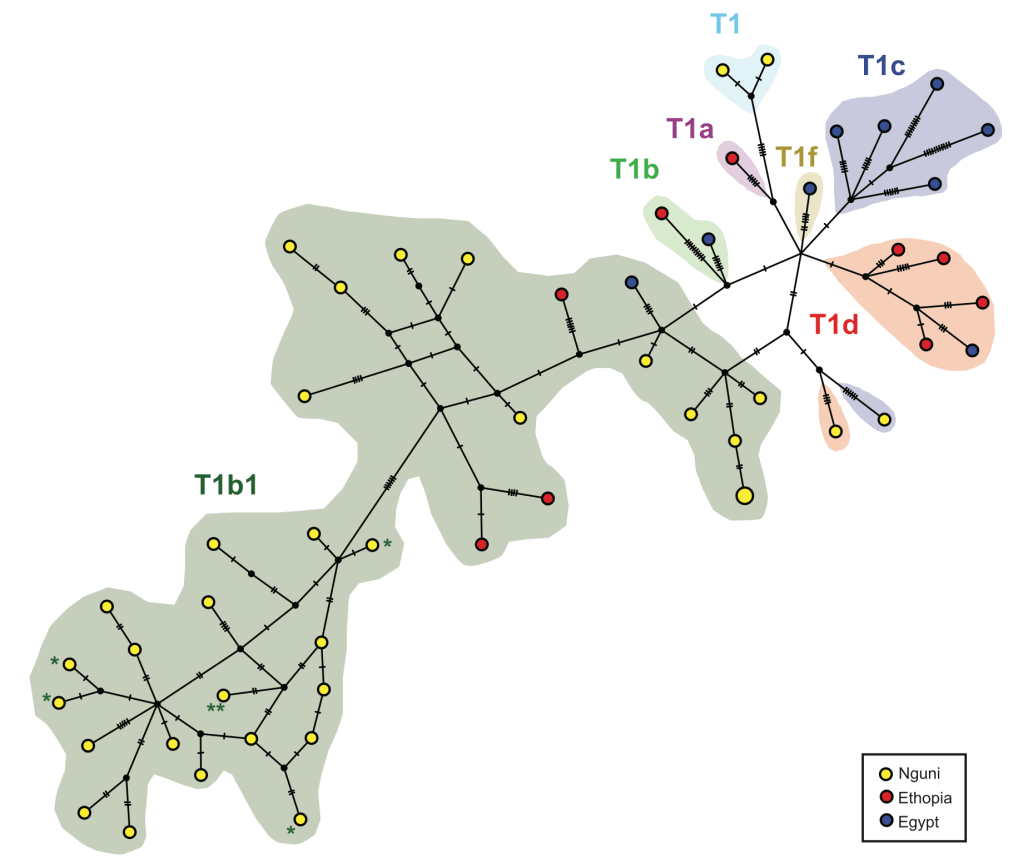
- For the modern sample, we want to examine the genomic effect of breeding strategies that introduced cattle from East and Northern Africa and Europe as a result of the rinderpest panzootic in the 1890s that killed most cattle in sub-Saharan Africa.
- We also want to explore whether herd management strategies, practices such as raiding, and Zulu engagement in Indian Ocean trade are evident in the maternal inheritance of cattle and the genetic diversity of herds dating to the Zulu Kingdom period.
Genetics Team

K. Ann Horsburgh, Genetics Team Lead
Ann is an Assistant Professor in Anthropology at Florida State University and a specialist in ancient mitochondrial DNA. As Director of the FSU Molecular Anthropology Research Group, her interests span anthropological issues in human ecology, human origins, migration, and culture contact. Her work on domesticated sheep and cattle from southern African sites has both challenged the misidentification of domesticated animals in faunal assemblages and thus far defined our expectations for precolonial sheep and cattle lineages from the southern third of the continent.
More on the work by Ann and her colleagues can be found here.
